PSTAT 100: Lecture 10
Estimation, Confidence Intervals, and Resampling
Department of Statistics and Applied Probability; UCSB
Summer Session A, 2025
\[ \newcommand{\Prob}{\mathbb{P}} \newcommand\R{\mathbb{R}} \newcommand{\N}{\mathbb{N}} \newcommand{\E}{\mathbb{E}} \newcommand{\Prob}{\mathbb{P}} \newcommand{\F}{\mathcal{F}} \newcommand{\1}{1\!\!1} \newcommand{\comp}[1]{#1^{\complement}} \newcommand{\Var}{\mathrm{Var}} \newcommand{\SD}{\mathrm{SD}} \newcommand{\vect}[1]{\vec{\boldsymbol{#1}}} \newcommand{\Cov}{\mathrm{Cov}} \newcommand{\iid}{\stackrel{\mathrm{i.i.d.}}{\sim}} \]
Recap: Statistical Inference
- Yesterday we talked about the general framework of statistical inference.
We have a population, governed by a set of population parameters that are unobserved (but that we’d like to make claims about).
To make claims about the population parameters, we take a sample.
We then use our sample to make inferences (i.e. claims) about the population parameters.
Recap: Statistical Inference
- A function of the random sample is an estimator
- This is a random quantity; “if I were to take a sample, …”
- The corresponding function of the observed instance (aka realization) of the sample is called an estimate
- This is a deterministic quantity; “given this particular sample I took, …”
Recap: Statistical Inference
The sampling distribution of an estimator is simply its distribution.
For example, we saw that the sampling distribution of the sample mean, assuming a normal population, is normal with mean equal to the population mean and variance equal to the population variance divided by the sample size.
- We further saw that, thanks to the Central Limit Theorem, this also holds if the population is not normal but the sample size is relatively large.
Let’s consider one more scenario: suppose \(Y_1, \cdots, Y_n\) represents an i.i.d. sample from the \(\mathcal{N}(\mu, \sigma^2)\) distribution where both µ and σ2 are unknown.
- Note that this differs from the situation we discussed yesterday, in which I had explicitly specified a value for σ2.
Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance
As mentioned yesterday, a natural estimator for the population variance is the sample variance, defined as \[ S_n^2 := \frac{1}{n - 1} \sum_{i=1}^{n} (Y_i - \overline{Y}_n)^2 \] and a natural estimator for the population standard deviation is just \(S_n := \sqrt{S_n^2}\).
Let’s take a look at the sampling distribution of \[ U_n := \frac{\sqrt{n}(\overline{Y}_n - \mu)}{S_n} \]
Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance

Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance
Hm… the bulk of this histogram looks normal, but there are some unusually extreme values that we wouldn’t expect to see if \(U_n\) were truly normally distributed.
Indeed, to check whether a set of values are normally distributed, we often generate a QQ-Plot in which we plot the quantiles of our data against the theoretical quantiles of a normal distribution.
- If the resulting graph is close to a perfect line, we know the quantiles match and our data is likely to be normally-distributed.
- Equivalently, if we see marked deviation from linearity in the tails of the plot, we have reason to believe our data is not normally-distributed.
Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance

Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance
As we see, there are significant deviations from linearity in the tails of the QQ-plot.
Hence, we conclude that \(U_n\) is not normally-distributed.
- That is, when we estimate the sample variance, the sampling distribution of the sample mean is no longer normal.
There is a theoretical result to back up this claim.
Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance
Sampling Distribution of Mean; Estimated Variance
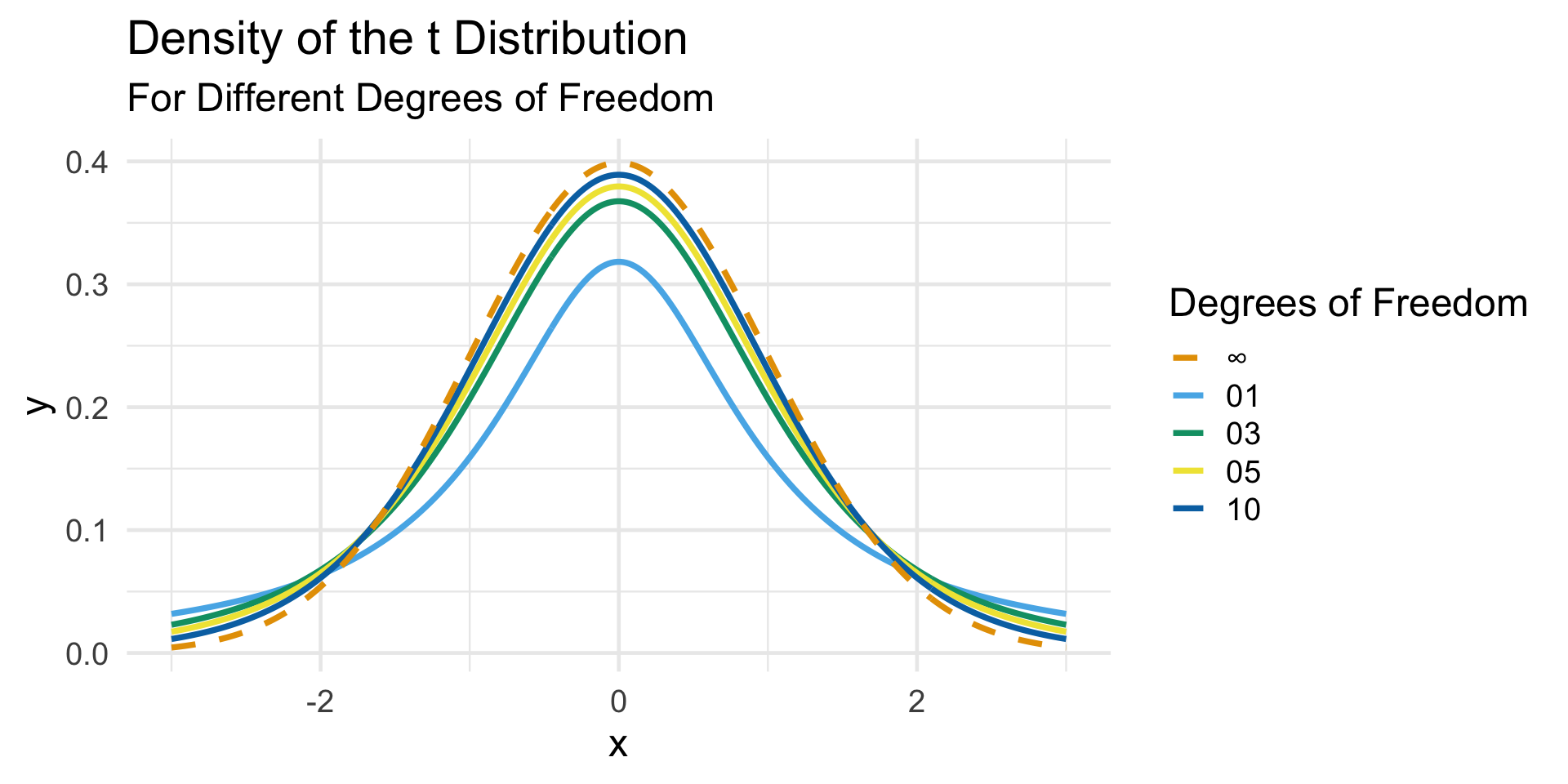
Given an i.i.d. sample (Y1, …, Yn) from a distribution with finite mean µ and finite variance σ2, \[ \frac{\sqrt{n}(\overline{Y}_n - \mu)}{S_n} \stackrel{\cdot}{\sim} t_{n - 1} \] where \(S_n := \sqrt{(n - 1)^{-1} \sum_{i=1}^{n} (Y_i - \overline{Y}_n)^2}\) and \(t_{n - 1}\) denotes the t-distribution with ν degrees of freedom.
- You will discuss the t-distribution further in PSTAT 120B. For our purposes, just note that the t-distribution looks a lot like a standard normal distribution but with wider tails.
Sampling Distributions
The t Distribution
Sampling Distributions
Sample Mean: Non-Normal Population; Unknown Variance

Confidence Intervals
Leadup
All of the estimators we considered thus far (sample mean, sample variance) are examples of point estimators; they reduce the sample to a single point.
In some cases, however, a point estimator may be too restrictive.
To borrow an analogy from OpenIntro Statistics (an introductory stats textbook I highly recommend!):
Using only a point estimate is like fishing in a murky lake with a spear. We can throw a spear where we saw a fish, but we will probably miss. On the other hand, if we toss a net in that area, we have a good chance of catching the fish. (pg. 181)
Confidence Intervals
The statistical analog of using a net is using a confidence interval.
Loosely speaking, a confidence interval is an interval that we believe, with some degree of certainty (called the coverage probability), covers the true value of the parameter of interest.
For example, we are 100% certain that the true average weight of all cats in the world is somewhere between 0 and ∞; therefore, a 100% confidence interval for µ (the true average weight of all cats in the world) is [0, ∞).
However, consider the interval [5, 20]. Are we 100% certain that the true average weight of all cats in the world is between 5 and 20 lbs? Probably not; so, the associated coverage probability of the interval [5, 20] is smaller than 100%.
Confidence Intervals
When constructing a Confidence Interval (CI), we often start with a coverage probability in mind first.
Some common coverage probabilities are: 90%, 95%, and 99%.
- This isn’t to say other coverage probabilities are never used; given domain knowledge, it may be desired to use a different coverage probability.
Suppose we wish to construct a p CI for the mean; i.e. given a population with (unknown) mean µ, we wish to construct a CI with coverage probability p (e.g. 0.95).
Confidence Intervals for the Mean
- It seems natural to start with \(\overline{Y}_n\), the sample mean. Indeed, we’ll first take our CI to be of the form \[ \overline{Y}_n \pm \mathrm{m.e.} \] for some margin of error \(\mathrm{m.e.}\).
- We can think of this as saying: “I think the sample mean is probably a good guess for the population mean. However, I acknowledge there is sampling variability, and I should include some padding in my estimate.”
- With this interpration, we see that the margin of error should depend on two things:
- The coverage probability p (higher p implies what about the margin of error?)
- The variance of \(\overline{Y}_n\) (to capture variability between samples).
Confidence Intervals for the Mean
So, let’s just take the margin of error to be the product of two terms: the variance of \(\overline{X}_n\), and a confidence coefficient c (a value related to our coverage probability p): \[ \overline{Y}_n \pm c \cdot \frac{\sigma}{\sqrt{n}} \]
So, all that’s left is to figure out what the confidence coefficient c should be.
Indeed, we should select it such that \[ \Prob\left( \overline{Y}_n - c \cdot \frac{\sigma}{\sqrt{n}} \leq \mu \leq \overline{Y}_n + c \cdot \frac{\sigma}{\sqrt{n}} \right) = p \]
Confidence Intervals for the Mean
Equivalently, \[ \Prob\left( -c \leq \frac{\sqrt{n}(\overline{Y}_n - \mu)}{\sigma} \leq c \right) = p \]
Hey- I know the sampling distribution of the middle quantity (provied we either have a normally-distributed population, or a large enough sample size for the CLT to kick in)!
So, our equivalent condition c must satisfy is \[ 2 \Phi(c) - 1 = p \ \implies \ c = \Phi^{-1}\left( \frac{1 + p}{2} \right) \]
Confidence Intervals for the Mean
CI for the Mean; Known Variance
Suppose (Y1, …, Yn) represents an i.i.d. sample from a distribution with finite mean µ and known variance σ2 < ∞. If either the Yi’s are known to be normally distributed or the sample size n is large (n ≥ 30), a p confidence interval for µ is given by \[ \overline{Y}_n \pm \Phi^{-1}\left( \frac{1 + p}{2} \right) \cdot \frac{\sigma}{\sqrt{n}}\]
- For example, the confidence coefficient associated with a 95% interval for the mean is given by
Confidence Intervals for the Mean
CI for the Mean; Unknown Variance
Suppose (Y1, …, Yn) represents an i.i.d. sample from a distribution with finite mean µ and unknown variance σ2 < ∞. If either the Yi’s are known to be normally distributed or the sample size n is large (n ≥ 30), a p confidence interval for µ is given by \[ \overline{Y}_n \pm F_{t_{n - 1}}^{-1} \left( \frac{1 + p}{2} \right) \cdot \frac{S_n}{\sqrt{n}}\] where \(F_{t_{n - 1}}^{-1}(\cdot)\) denotes the inverse CDF of the tn-1 distribution.
- For example, the confidence coefficient associated with a 95% interval for the mean, given a sample size of 32, is given by
Your Turn!
Your Turn!
A consultant from the EPA (Environmental Protection Agency) is interested in estimating the average CO2 emissions among all US households. To that end, they collect a sample of 35 US households; the average emissions of these 35 households is 9.13 mt/yr and the standard deviation of these emissions is 2.43 mt/yr. Construct a 90% confidence interval for the true average CO2 emissions among all US households using the consultant’s data.
Caution
You’ll need a computer for this one!
05:00
Resampling Methods
Leadup
- Much of our discussion yesterday and today has been simulation-based: for instance, yesterday we repeatedly sampled from the Exponential distribution and constructed the empirical sampling distribution for the sample mean.
- Furthermore, in our simulations, we sampled directly from the population.
- What happens if we don’t have access to the population distribution?
- For example, suppose we have 100 cat weights, but we don’t want to necessarily assume that the weight of a randomly-selected cat follows the normal distribution.
Resampling Methods
Here is an idea: what if we treat this sample as the population itself?
- If we do, we can simply generate as many samples (with replacement) as we like and still construct empirical sampling distributions for estimators using the procedure we implemented yesterday.
This is the idea behind the bootstrap, which itself belongs to a class of techniques known as resampling methods.
Let’s run through an example together.
Imagine we have a vector of 100 cat weights, stored in a variable called
cat_wts.
Bootstrapping: Example
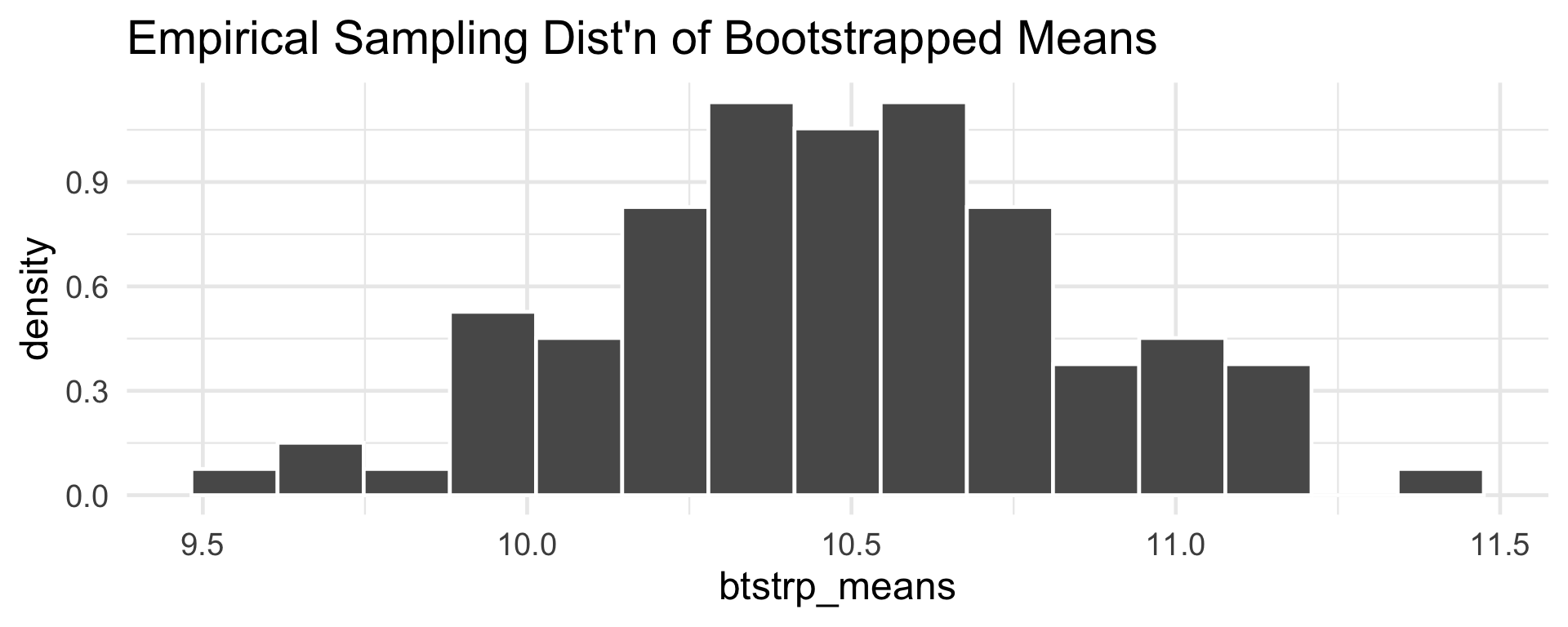
- We can take a resample of size 100 (it’s customary to take resamples of the same size as the original sample):
- Here’s how the mean of our resample compares with the mean of our sample:
Bootstrapping: Example
- If we imagine repeating this resampling procedure, we can construct an approximation to the sampling distribution of the sample mean.
Bootstrapping: Example
Since this data was simulated, we can actually assess how well the bootstrap is doing.
Specifically (even though I did not tell you this before), the
cat_wtsvector was sampled from the \(\mathcal{N}(10.5, \ 3.2^2)\) distribution.So, let’s compare our bootstrapped sample means against a set of sample means drawn directly from the population distribution.
Code
Min. 1st Qu. Median Mean 3rd Qu. Max.
9.285 10.276 10.493 10.499 10.711 11.482 Min. 1st Qu. Median Mean 3rd Qu. Max.
9.549 10.237 10.482 10.482 10.704 11.408 Bootstrapping: Example
Code
data.frame(`From Pop.` = sm_from_pop,
`Bootstrapped` = btstrp_means,
check.names = F) %>%
melt(variable.name = "type") %>%
ggplot(aes(x = type, y = value)) +
geom_boxplot(fill = "#dce7f7", staplewidth = 0.25,
outlier.size = 2) + theme_minimal(base_size = 18) +
ggtitle("Boxplot of Means",
subtitle = "Sampled from Population, and Bootstrapping")
Bootstrapping CIs
What this means is that we can actually construct confidence intervals using bootstrapping.
There are actually many different flavors of bootstrapped CIs; we’ll only discuss one in this class.
Specifically, let’s go back to how I defined a confidence interval: an interval that we believe, with coverage probability, covers the true value of the parameter of interest.
An equivalent way of interpreting a p CI is: if we were to repeat the mechanism used to generate the CI a large number of times, we would expect (on average) (p×100)% of the resulting CIs to cover the true value of µ.
CI: Interpretation
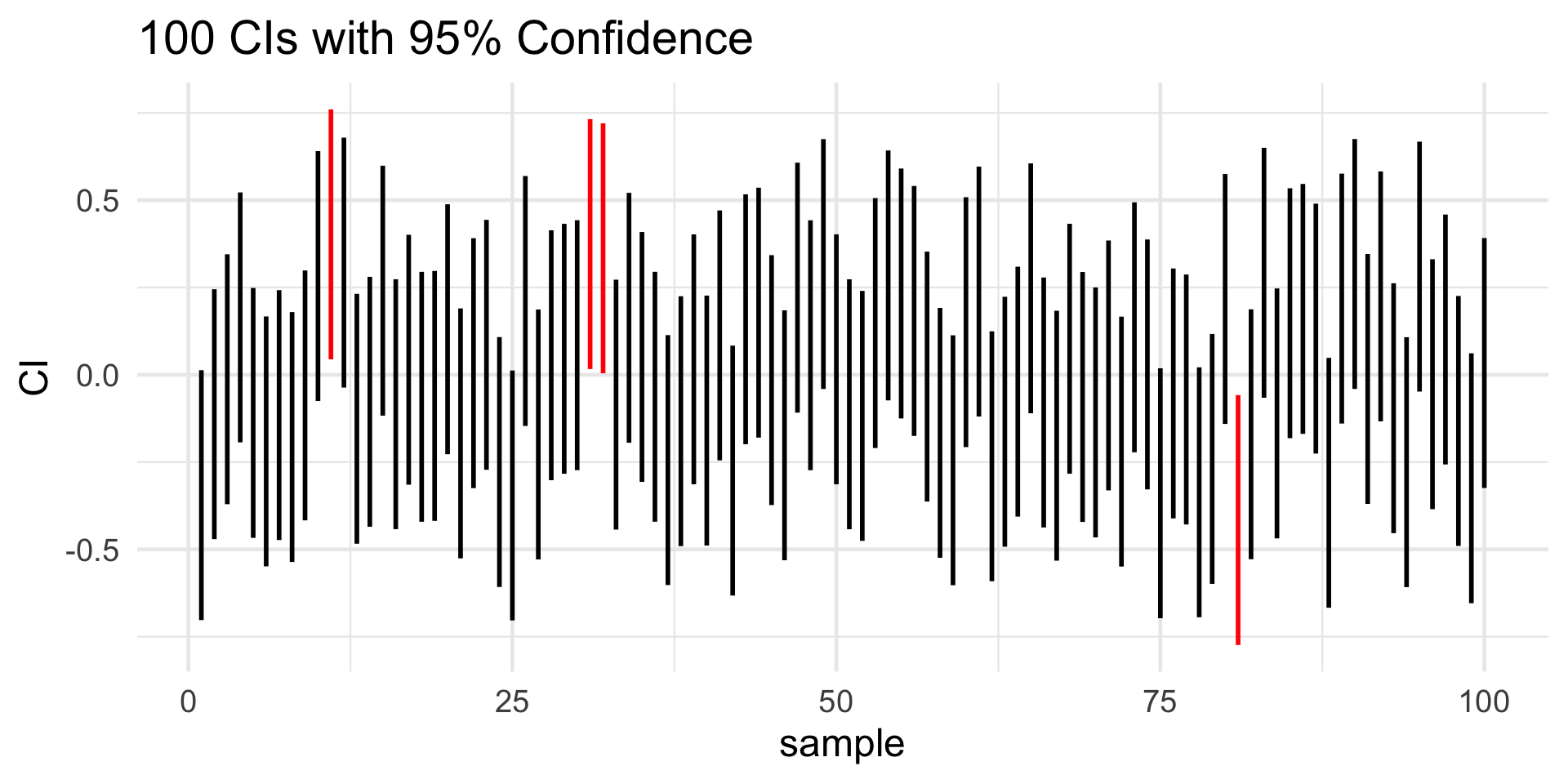
Bootstrapping CIs
- “If we were to repeat […] large number of times”; isn’t that exactly what we do in the bootstrap?
- Yup!
- So, here’s a way we can use the Bootstrap to obtain a p CI:
- Generate a large number of bootstrapped estimators
- Use the (1 - p)/2 and (1 + p)/2 quantiles of the bootstraped sampling distribution
- This has the benefit of being “distribution free” (sometimes called nonparametric), and can be used to construct confidence intervals for a wide array of parameters (not just the mean).
PSTAT 100 - Data Science: Concepts and Analysis, Summer 2025 with Ethan P. Marzban